missing translation for 'onlineSavingsMsg'
Learn More
Learn More
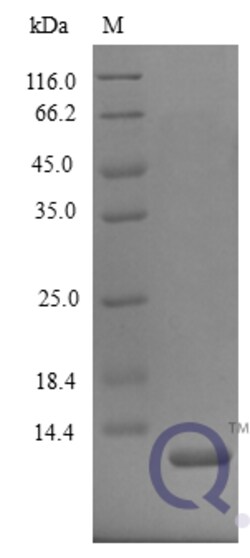
enQuireBio™ Recombinant Mouse CXCL2 / MIP-2 Protein
Click to view available options
Quantity:
100 μg
20 μg
250 μg
5 μg
Unit Size:
100µg
20µg
250µg
5µg
Specifications
Specifications
| Accession Number | NP_033166.1 |
| Gene ID (Entrez) | 20310 |
| Name | CXCL2 / MIP-2 Protein |
| Quantity | 5 μg |
| Regulatory Status | Research Use Only |
| Endotoxin Concentration | Less than 1.0 EU/ug as determined by LAL method. |
| Gene Symbol | CXCL2 |
| Protein Length | Partial (see sequence information for more details). |
| Biological Activity | The biological activity determined by a chemotaxis bioassay using human neutrophils is in a concentration range of 1.0-10 ng/ml. |
| Product Type | Recombinant Protein |
| Show More |
Product Content Correction
Your input is important to us. Please complete this form to provide feedback related to the content on this product.
Product Title
Spot an opportunity for improvement?Share a Content Correction